Scientists at the Frederick National Laboratory for Cancer Research and investigators across the National Institutes of Health (NIH) have developed a highly specific serology test to determine whether a person has antibodies to SARS-CoV-2, a vital tool to understanding the spread of infection from the virus that causes COVID-19.
The collaboration started in March to optimize and standardize an antibody test to be deployed in the NIH serosurvey study of 10,000 people to evaluate the spread of the pandemic. The findings and standardization process was published online May 25 in a preprint, a version of the paper before it is examined by peer scientists.
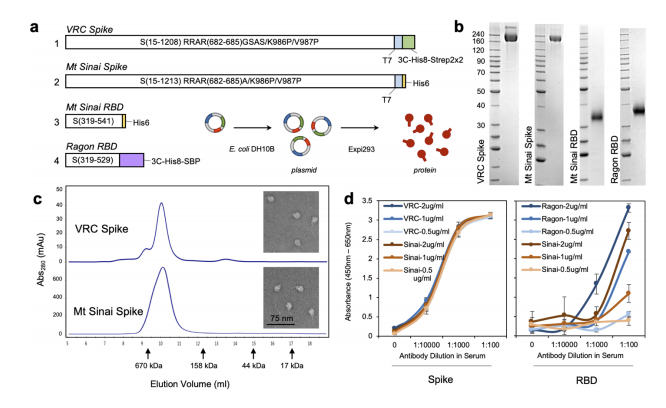
To develop the serology test, the FNL’s Protein Expression Laboratory generated initial SARS-CoV-2 protein samples to see which would most effectively determine the presence of antibodies to the virus. The Protein Expression Laboratory, led by Dominic Esposito, Ph.D., is part of the National Cancer Institute’s RAS Initiative, headquartered at FNL.
Esposito said the research identified optimal forms of the CoV-2 spike protein and spike receptor-binding domain (RBD) protein to produce a serology test with maximum sensitivity. He said by using both proteins, they managed to narrow the test for antibodies.
“If you only look at one protein or the other, you get more false positives or negatives,” Esposito said. “You have to fit into that box that is both positive for RBD and spike for it to be a true positive.”
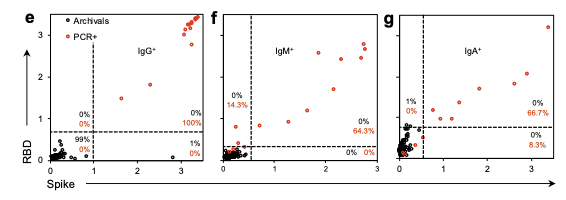
The preliminary findings suggest the serology test has well above 99 percent specificity. The study was conducted by researchers from the National Institute of Biomedical Imaging and Bioengineering, the National Center for Advancing Translational Sciences, the National Institute of Allergy and Infectious Diseases and the FNL.
To validate the process, the team tested blood samples from patients with a confirmed COVID-19 diagnosis, from a blood drive conducted in a community with confirmed cases and from people who had been exposed to the virus. Results were compared to blood samples from healthy volunteers in a NIH study prior to 2019. These samples were used as negative controls for SARS-CoV-2 to define the threshold for seropositivity, which means the patient’s blood shows the presence of an antibody for a given pathogen.
The team also compared SARS-CoV-2 antibodies to antibodies to other coronaviruses such as MERS, SARS1 and viruses that cause the common cold. The purpose was to tighten parameters for the serology test by focusing on specific antibodies unique to SARS-CoV-2, Esposito said. These antibodies will appear whether a patient had symptoms or no symptoms at all, helping to determine the spread of infection among the population.
The serology test developed at FNL is part of the National Cancer Institute’s broader effort to develop a program to develop, validate, improve, and implement serological testing and associated technologies in response to the COVID-19 pandemic. Widespread availability of serology tests – or antibody tests – is critical to understanding who has been infected with SARS-CoV-2 and may be immune to future infection, though questions remain on whether having antibodies means a person is protected against reinfection and how long immunity lasts.
Media Inquiries
Mary Ellen Hackett
Manager, Communications Office
301-401-8670
