
Researchers trained deep learning models to predict the aggressiveness of rare soft tissue tumors, known as rhabdomyosarcomas, based on images of thin slices of tumors obtained during biopsies.
By training computer models to assess pathology images of rhabdomyosarcoma tumors, researchers hope to help doctors better tailor treatments to patients.
Researchers at the Frederick National Laboratory for Cancer Research and the National Cancer Institute developed an artificial intelligence-based system that can anticipate the severity of cases of a childhood cancer using histopathologic images of tumors. With more development, these computer models, which are in the early stages of development, could help doctors select the course of therapy best suited to each patient.
A cancer that arises in soft tissue, such as muscle, rhabdomyosarcoma predominantly affects children and young adults. While some therapies targeting specific genetic mutations are being studied, treatment regimens typically include chemotherapy, radiation, and surgery. Doctors select the course of treatment based on how aggressive tumors appear, an assessment based on factors that include the cancer’s location in the body and whether it has spread. The outlook for those with this disease, known as rhabdomyosarcoma, can vary considerably.
In a study published in January 2023 in Clinical Cancer Research, researchers found their AI-based system could assess the risk that patients’ disease would progress at least as well as the conventional approach. The team also found that, from these histopathologic images, their models could determine if tumors possessed certain genetic mutations.
The variety of artificial intelligence they used, known as deep learning, has become the new go-to approach for analyzing medical images, says study co-author Hyun Jung, a bioinformatics manager at FNL who specializes in image processing and computer vision specialist.
However, these models require a great deal of data from patients to sufficiently train and validate — a challenge for rare diseases like rhabdomyosarcoma, he notes.
“With enough additional development, deep learning models like ours could be integrated into pathologists’ workflow in the near future, helping them to make more precise predictions for these patients,” Jung says.
Assessing tumors
Although the use of genetic sequencing to detect cancer-driving mutations is increasing, doctors do not routinely order these tests for rhabdomyosarcoma. More and more, however, assessments are including one of many genetic factors known to influence this malignancy’s behavior: the fusion of two genes, FOXO1 and PAX3 or 7, which is associated with an often more aggressive form, known as alveolar rhabdomyosarcoma.
Credit: Javed Khan
In tumors that lack these aberrant gene fusions, typically those called embryonal rhabdomyosarcoma, many other mutations appear. Researchers linked some of these other changes to more aggressive clinical disease. Yet, while doctors take gene fusion, or the presence of the embryonal versus alveolar variety of the disease, into account when assessing risk, they generally do not look for these other mutations.
As an alternative, researchers have begun experimenting with deep learning to assess tumors. In previous efforts, other groups distinguished embryonal and alveolar tumors, and assessed the risk of progression using deep learning models. This most recent study, however, is the first to train models to recognize genetic mutations.
The advantage of deep learning
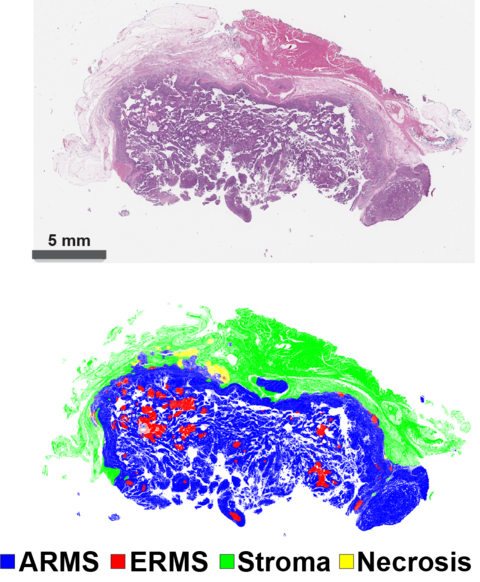
To develop their models, the team relied on images of thin slices of tumors obtained from biopsies, like those pathologists traditionally analyzed under a microscope. Jung and his colleagues used these images to train a model to distinguish the types of tissues — supportive, cancerous, or just dead — contained in the biopsy slides, just as a pathologist would.
They developed five other models that focused only on the tumor tissue. By reducing the images of the cancerous tissue down into small groups of pixels (digital picture elements), these models are trained to make predictions based on characteristics that researchers themselves cannot identify.
“One of the big advantages of using deep learning is that it extracts those features by itself, we don’t have to give it any specific instructions,” Jung says.
To assess the aggressiveness of the disease, the researchers gave one of the models data on how patients fared. With this additional information, it learned to classify tumors as doctors would, into three categories: high, intermediate, or low risk of progression or mortality.
When they tested it, they found the model performed comparably to the conventional assessment system, except that it better separated high and intermediate risk cases.
In a similar manner, they trained other models to recognize tumors that possessed fused genes, or other mutations. These included errors in genes called MYOD1 and TP53, which are linked to aggressive cases, and within a molecular pathway known as RAS.
Toward tailored therapy
Mutation-detecting models like these could potentially flag cases that merit genetic sequencing, according to corresponding author Javed Khan, a translational physician and oncologist at NCI. Meanwhile, the risk assessment model could potentially catch tumors that might behave aggressively even though they don’t possess any high-risk mutations, he says.
Khan notes that the team developed these models based on 321 patients, a relatively small number. They plan is to improve them using data from over 1,500 more cases of rhabdomyosarcoma, he says.
“We are at an early stage,” Khan says. “With more data and more training, these deep learning models will get very powerful, not just for this type of cancer, but for many cancers.”
Media Inquiries
Mary Ellen Hackett
Manager, Communications Office
301-401-8670
